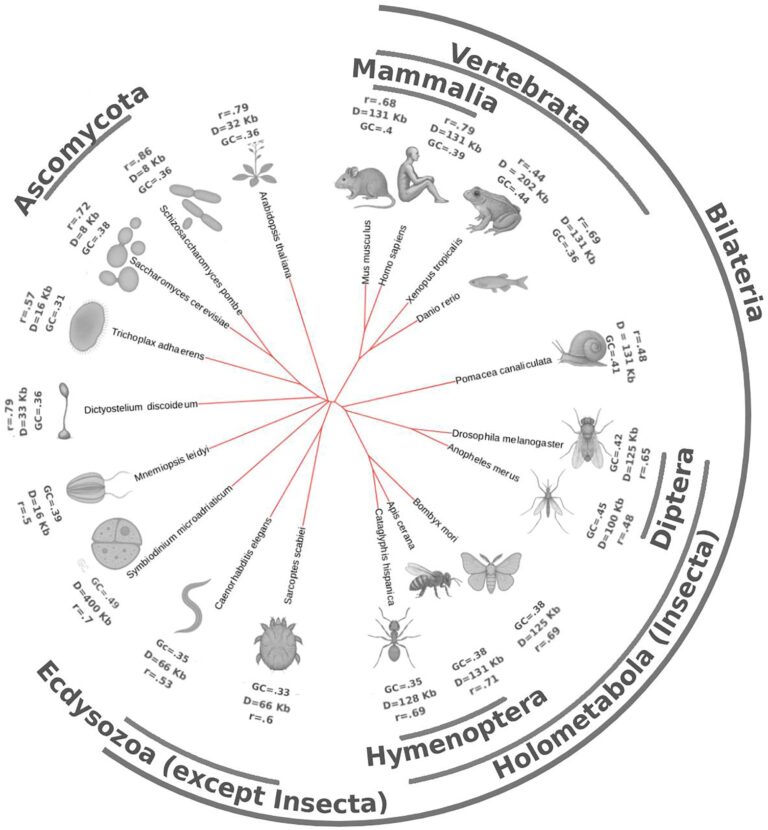
Chromatin-based tree of life
In the new work published today in NAR, Aleksei Shkolikov and I explore what we can see in Hi-C — with convolutional autoencoder — across the tree of life in 22 species, ranging from amoeba and dinoflagellate to vertebrates and insects. With our approach called Chimaera, we learn the rules