Does chromatin organization shape immune responses? Our recent study with Nick Adams led by Reizis Lab and Leonid Mirny explores how cohesin, a key player in chromatin loop extrusion, controls the chromatin architecture essential for conventional dendritic cells (cDC).
DCs bridge innate and adaptive immunity. Surprisingly, strictly required factor for DCs to function is cohesin. Without it, cDCs fail to differentiate fully, but more critically, lose ability to cross-present antigens and secrete IL-12, a key cytokine for immune defense.
Cohesin organizes chromosomes, but not only. In DCs, the response to the antigens is impaired without cohein! We figured out that genes that fail to get activated are those far (>100 Kb) from the nearest enhancers and more enriched in Polycomb before activation.
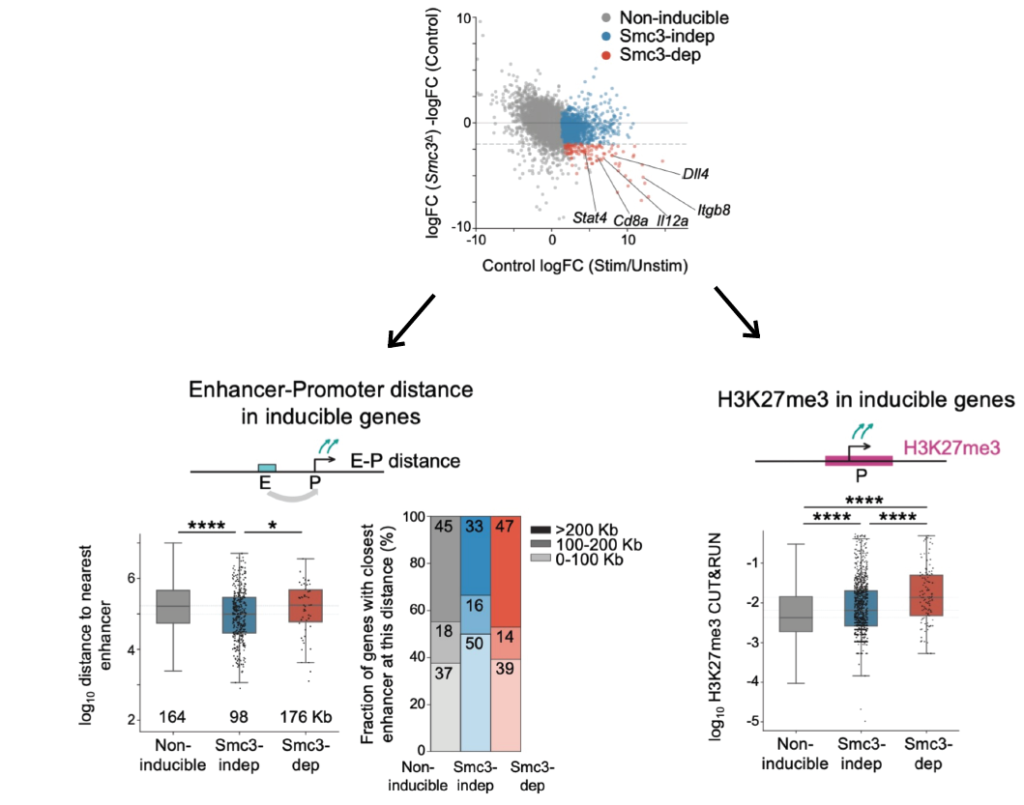
It aligns well with recent studies showing the role of cohesin in shaping promoter-enhancer interactions and counteracting the Polycomb repression!
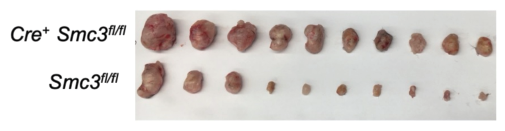
Mice deficient in cohesin show impaired IL-12 production and weakened anti-tumor responses, suggesting that cohesin is crucial for activating immune cells and proper cytokine signaling. This could have implications for cancer therapies! My favorite figure by Nick:
Cohesin complex doesn’t work alone in shaping the dendritic cells function. Transcription factor IRF8 ensures the proper chromatin organization, most significantly directing the compartmental switches of genes active in DCs and their activation.
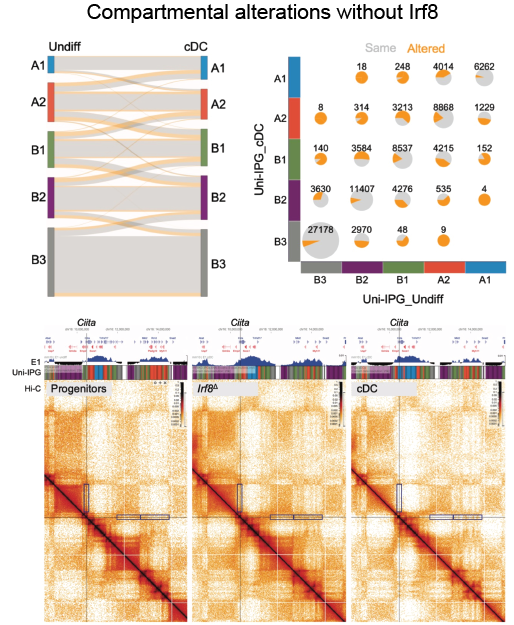
Coheins-Irf8 dynamic duo optimizes DC function by managing chromatin loops and compartmentalization.
The big takeaway: cohesin isn’t just about genome structure—it’s an essential regulator of immune function, influencing how DCs fight pathogens and tumors.
Read more and see annotations to the figures in our bioRxiv preprint!