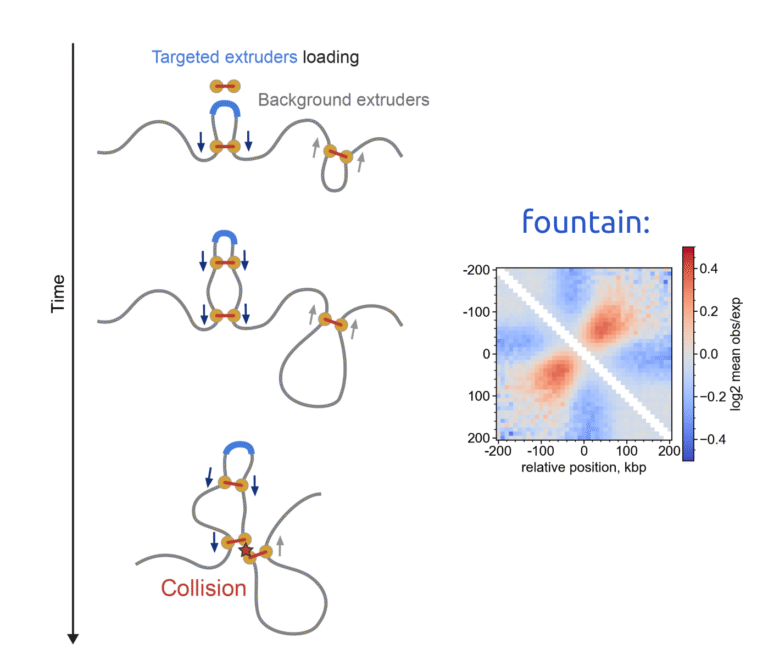
In this update, I show that chromatin “fountains” form at enhancers not only in early zebrafish embryos but also during mitotic exit in mouse embryonic stem cells (mESCs). This is exciting because fountains appear to be a universal early feature of genome folding and likely reflect cohesin loading at enhancers.
My summary below focuses on the polymer-simulation side. To reproduce the characteristic fountain signature in Hi-C/Micro-C, you need (i) targeted loading of loop extruders (e.g., cohesin) at specific genomic sites—likely enhancers—and (ii) background obstacles that impede cohesin’s reeling. Together, these ingredients generate the fountain-like enrichment pattern in Hi-C.
The original preprint appeared on bioRxiv in 2023, and we’re now sharing a revised version. More details:
https://www.biorxiv.org/content/10.1101/2023.07.15.549120v2.full