I’m pleased to share the results of a project I started during my graduate studies, now beautifully completed by my colleagues.
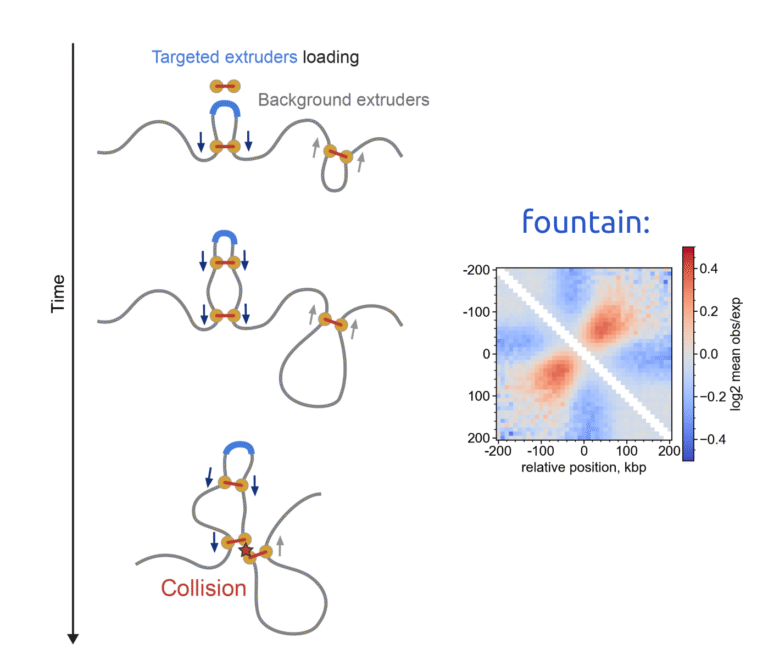
The study applies a sophisticated method to construct joint embeddings of DNA sequences and Hi-C chromatin organization maps within a shared space. This framework allows for the exploration of chromatin structures with distinct functional characteristics, such as insulation domains, loops, and fountains. Intriguingly, it also enables us to identify DNA sequences associated with specific structural features and even modify chromatin maps by ‘adding’ these elements.
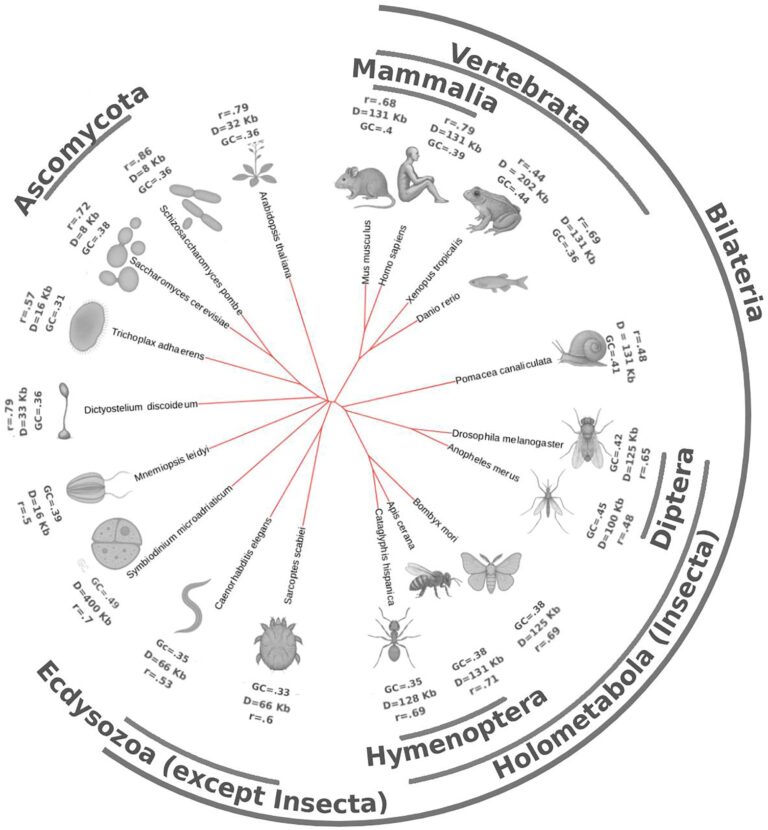
One particularly striking result is the construction of a ‘tree of life’ for chromatin spatial organization. While it bears some similarity to traditional phylogenetic trees based on DNA sequences, there are notable differences that reflect significant evolutionary shifts in mechanisms of chromatin folding.