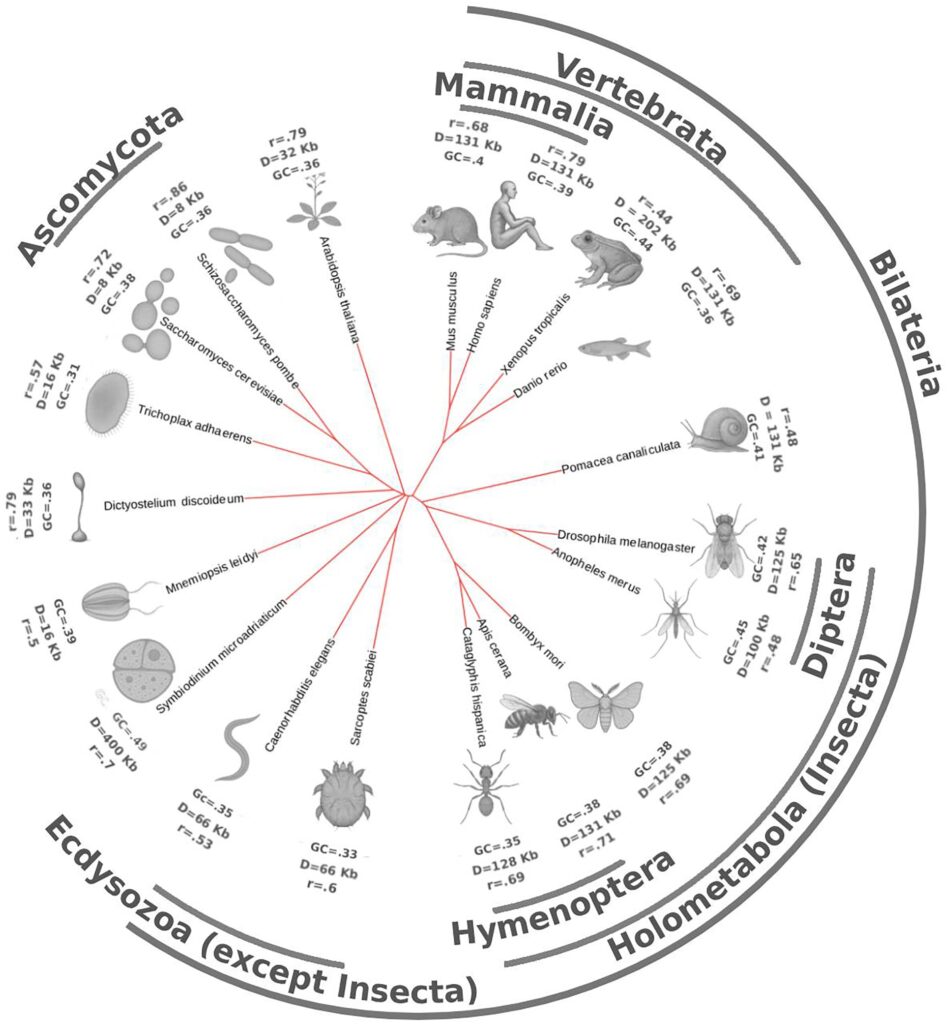
In the new work published today in NAR, Aleksei Shkolikov and I explore what we can see in Hi-C — with convolutional autoencoder — across the tree of life in 22 species, ranging from amoeba and dinoflagellate to vertebrates and insects.
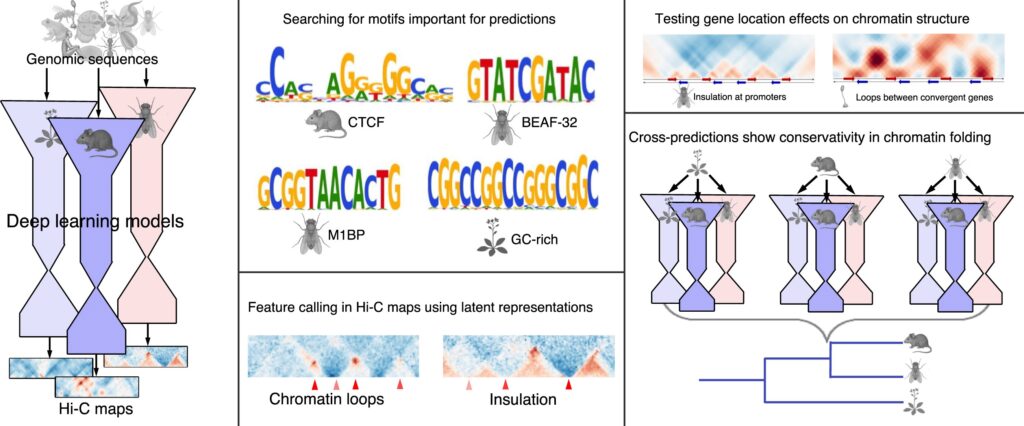
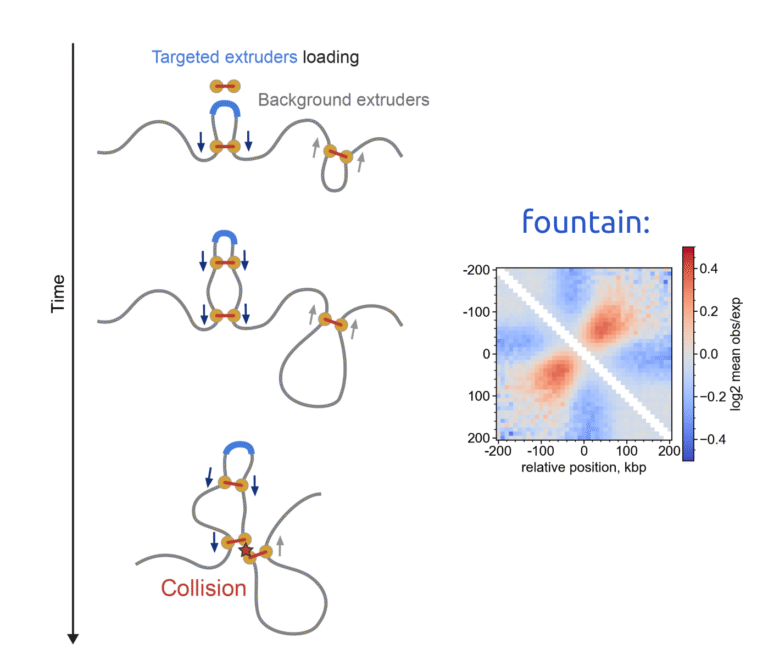
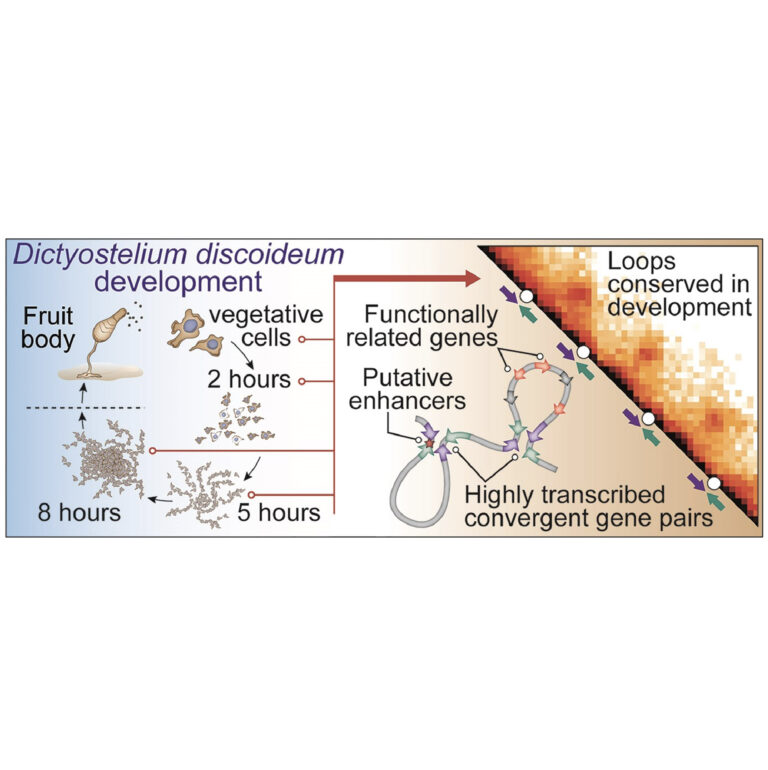
With our approach called Chimaera, we learn the rules of 3D genome folding and their connection to the underlying DNA sequence determinants. This approach allows for interesting gameplay to unfold: calling chromatin patterns (such as loops, dots, TADs and fountains), inspecting the presence of these patterns in unusual scenarios (embryogenesis and mitosis), and detecting DNA motifs associated with these patterns.
The most fascinating result (as for me) is the chromatin rules-based tree of life that we present at the end of this massive work. Our (mammalian) chromatin folds quite differently from the rest of the life forms!
There are many more cool results in this paper — and mysterious DNA motifs associated with insulation patterns in Arabidopsis and Drosophila. Check it out: https://doi.org/10.1093/nar/gkaf1516
If you got curious and want to get involved with projects like this one — drop me a message at agalitzina at gmail.com